In this script, I attempt to use calmr’s implementation of SOCR to reproduce some of the figures in Witnauer et al. (2012).
The fixed parameters
In Witnauer et al (2012), they set the alphas of cues, contexts, and the US to 0.35, 0.15, and 0.50, respectively. The rate at which associations weaken (k1, here, omega) was set to be 0.22. The weight with which comparison stimuli compete (k2, here, gamma) was set to 0.56. The rate at which the operator switch is learned (k3, here, tau) was set to 0.46. Finally, the strength with which present stimuli are activated (k4, here, rho; see Eq. 4 in their paper) was set to 1.60.
Acquisition
mod <- "SM2007"
des <- data.frame(
Group = "Acquisition",
P1 = "100(ctxa)A>(US)/300(ctxa)",
R1 = FALSE,
P2 = "1#(ctxa)A/1#(ctxb)A",
R2 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .15, .5)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
pars## $alphas
## A ctxa ctxb US
## 0.35 0.15 0.15 0.50
##
## $lambdas
## A ctxa ctxb US
## 1 1 1 1
##
## $omegas
## A ctxa ctxb US
## 0.22 0.22 0.22 0.22
##
## $rhos
## A ctxa ctxb US
## 1.6 1.6 1.6 1.6
##
## $gammas
## A ctxa ctxb US
## 0.56 0.56 0.56 0.56
##
## $taus
## A ctxa ctxb US
## 0.46 0.46 0.46 0.46
##
## $order
## [1] 1
m <- run_experiment(des,
model = mod,
parameters = pars
)
# Fig 2 A
results(m)$relacts %>%
filter(s2 == "US" & trial_type == "(ctxa)A>(US)" & phase == "P1") %>%
mutate(trial = ceiling(trial / block_size)) %>%
group_by(trial) %>%
summarise(value = sum(value)) %>%
ggplot(aes(x = trial, y = value)) +
geom_line() +
geom_point()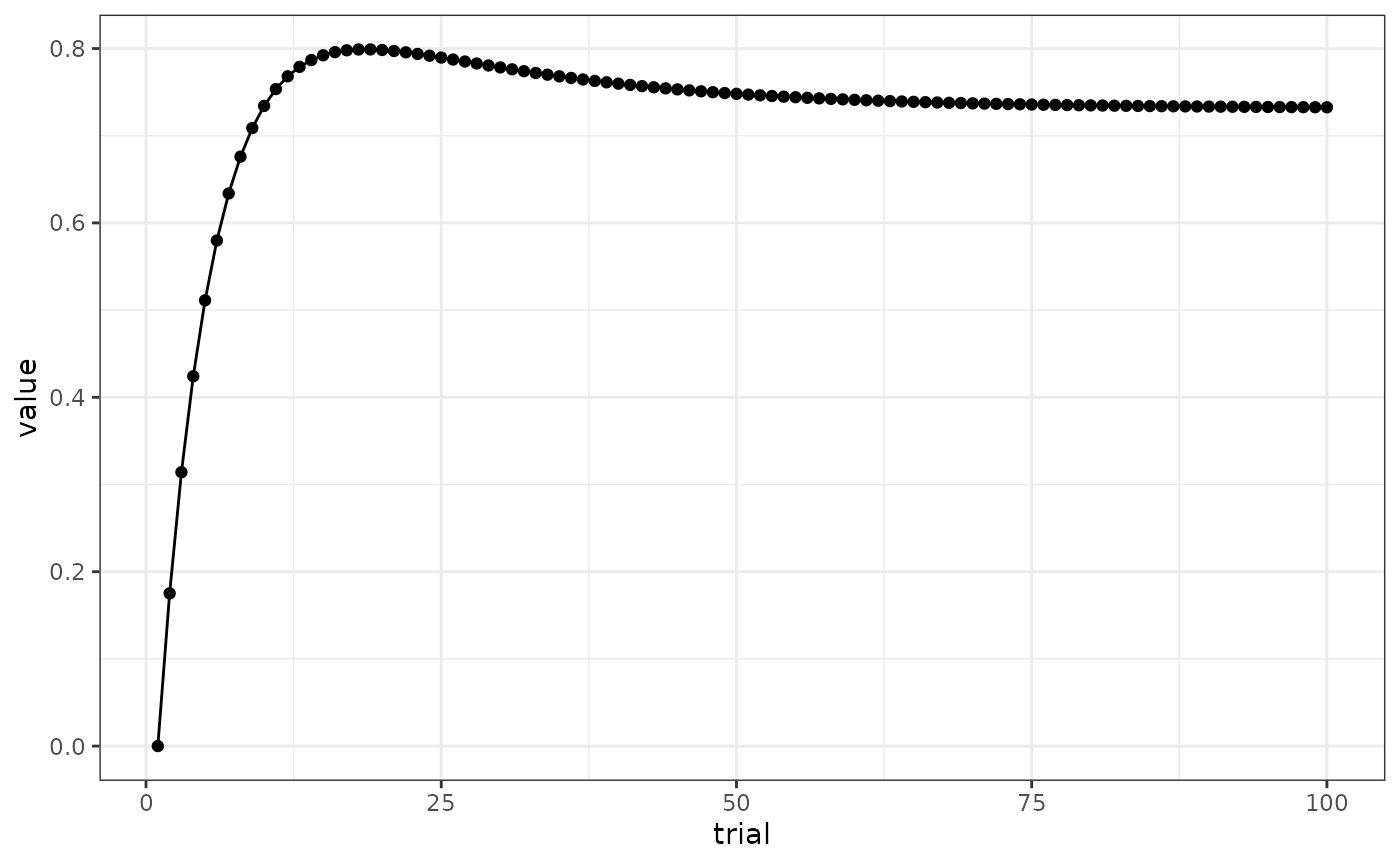
# overall responding levels are higher for calmr
# Fig 2 B
results(m)$relacts %>%
filter(s2 == "US" & phase == "P2") %>%
group_by(trial_type) %>%
summarise(value = sum(value)) %>%
mutate(trial_type = ifelse(grepl("b", trial_type),
"Neutral Ctx", "Acquisition Ctx"
)) %>%
ggplot(aes(x = trial_type, y = value)) +
geom_col()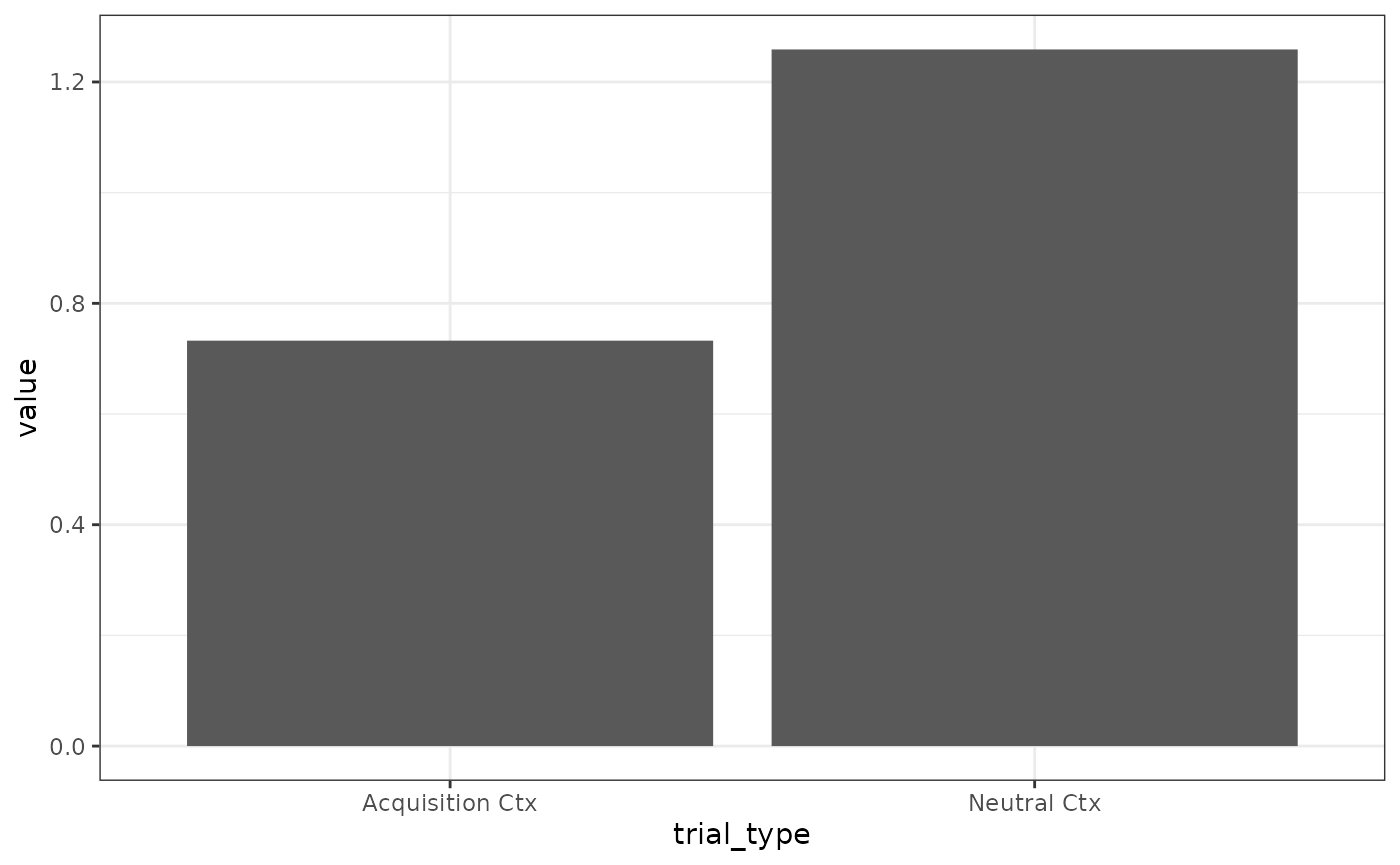
# overall responding levels are higher for calmrExtinction
des <- data.frame(
Group = "Extinction",
P1 = "10(ctxa)A>(US)/30(ctxa)",
R1 = FALSE,
P2 = "30(ctxb)A/90(ctxb)",
R2 = FALSE,
P3 = "1#(ctxa)A/1#(ctxb)A",
R3 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .15, .5)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
pars## $alphas
## A ctxa ctxb US
## 0.35 0.15 0.15 0.50
##
## $lambdas
## A ctxa ctxb US
## 1 1 1 1
##
## $omegas
## A ctxa ctxb US
## 0.22 0.22 0.22 0.22
##
## $rhos
## A ctxa ctxb US
## 1.6 1.6 1.6 1.6
##
## $gammas
## A ctxa ctxb US
## 0.56 0.56 0.56 0.56
##
## $taus
## A ctxa ctxb US
## 0.46 0.46 0.46 0.46
##
## $order
## [1] 1
m <- run_experiment(des,
model = mod,
parameters = pars
)
# Fig 4 A
results(m)$relacts %>%
filter(s2 == "US" & trial_type == "(ctxb)A" & phase == "P2") %>%
mutate(trial = ceiling(trial / block_size)) %>%
group_by(trial) %>%
summarise(value = sum(value)) %>%
ggplot(aes(x = trial, y = value)) +
geom_line() +
geom_point()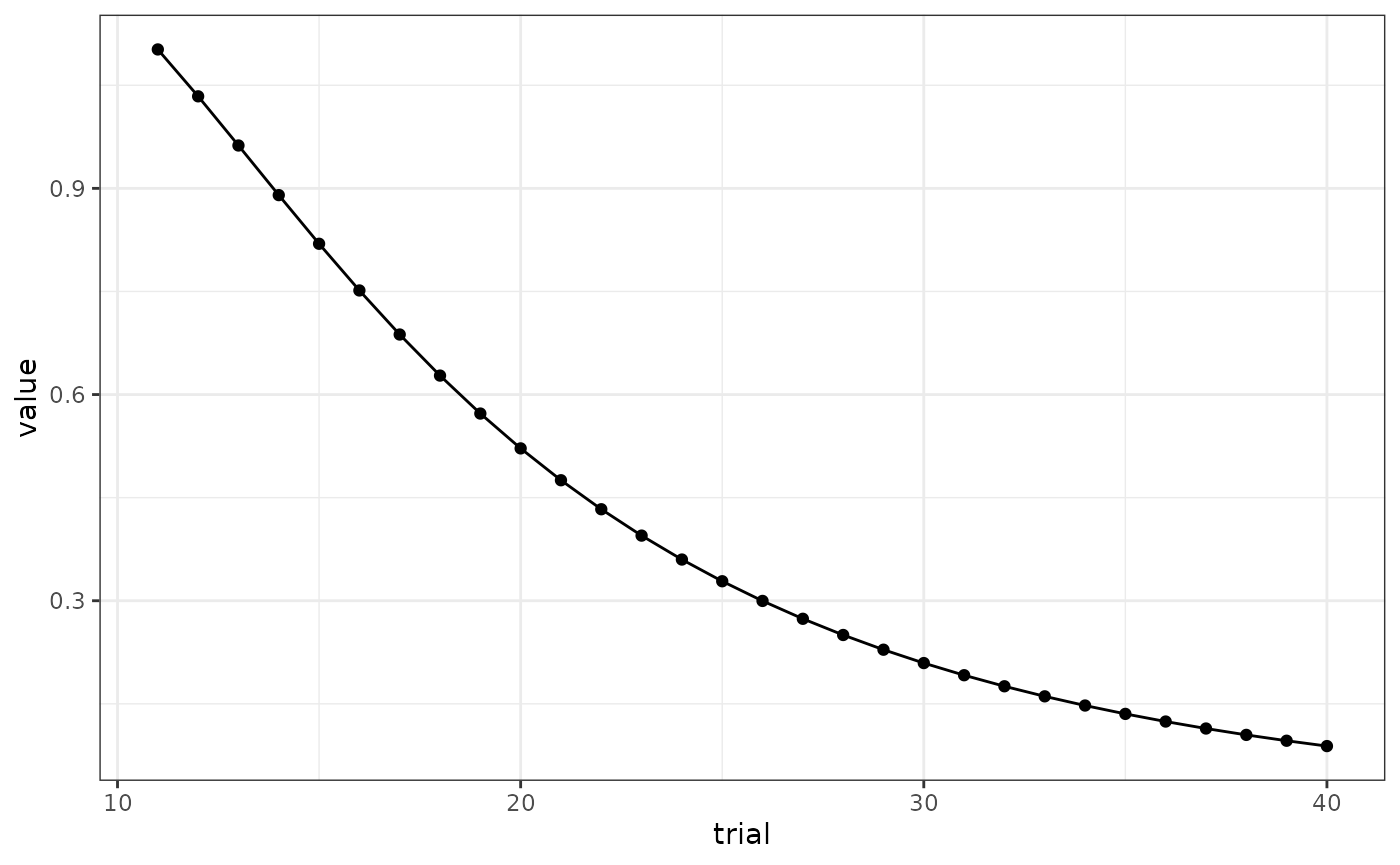
# responding at the beginning of extinction is higher
results(m)$relacts %>%
filter(s2 == "US" & phase == "P3") %>%
group_by(trial_type) %>%
summarise(value = sum(value)) %>%
mutate(trial_type = ifelse(grepl("b", trial_type),
"Extinction Ctx", "Acquisition Ctx"
)) %>%
ggplot(aes(x = trial_type, y = value)) +
geom_col() +
coord_cartesian(ylim = c(0, .4))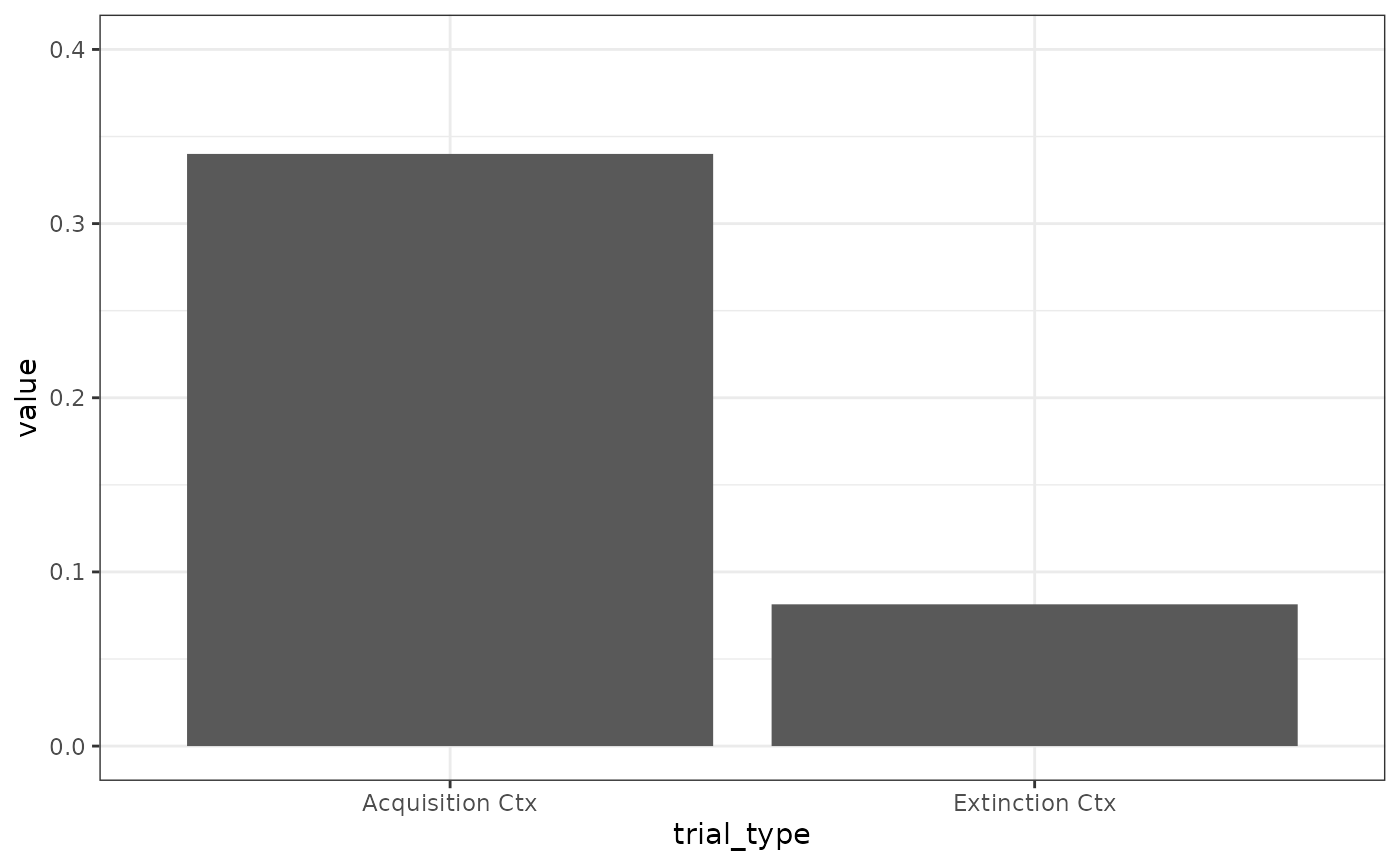
# remarkably similarSecond order conditioning/Conditioned inhibition
des <- data.frame(
Group = c("Few", "Many"),
P1 = c(
"10(ctx)A>(US)/10(ctx)AX/10(ctx)B>(US)/90(ctx)",
"100(ctx)A>(US)/100(ctx)AX/100(ctx)B>(US)/900(ctx)"
),
R1 = FALSE,
P2 = "1#(ctx)X/1#(ctx)BX",
R2 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .35, .35, .5)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
m <- run_experiment(des,
model = mod, parameters = pars
)
# Fig 6
results(m)$relacts %>%
filter(s2 == "US" & phase == "P2") %>%
group_by(group, trial_type) %>%
summarise(value = sum(value)) %>%
mutate(trial_type = ifelse(grepl("B", trial_type),
"Summation", "Elemental"
)) %>%
ggplot(aes(x = group, y = value, fill = trial_type)) +
geom_col(position = position_dodge())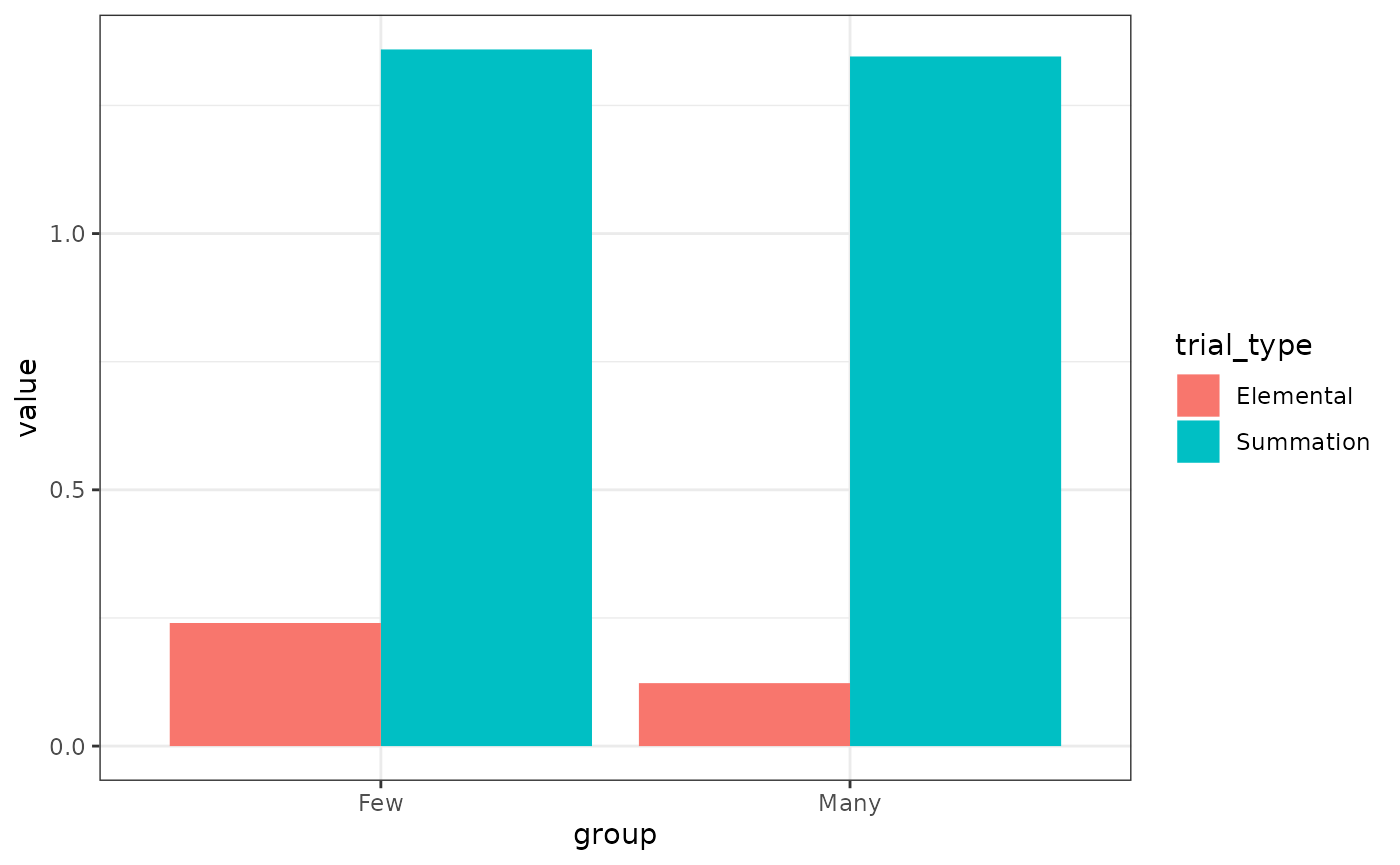
# pretty close, but scales are a little bit offBlocking
des <- data.frame(
Group = c(
"Elemental", "Overshadowing",
"Blocking", "Recovery From Blocking"
),
P1 = "10(ctx)A>(US)/180(ctx)",
R1 = FALSE,
P2 = c(
"5(ctx)X>(US)/90(ctx)",
"5(ctx)BX>(US)/90(ctx)",
"5(ctx)AX>(US)/90(ctx)",
"5(ctx)AX>(US)/90(ctx)"
),
R2 = FALSE,
P3 = c(
"", "", "", # nothing for the first three grups
"5(ctx)A/90(ctx)"
),
R3 = FALSE,
P4 = c("1#(ctx)X"),
R4 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .35, .35, .5)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
m <- run_experiment(des,
model = mod, parameters = pars
)
# Fig 10
results(m)$relacts %>%
filter(s2 == "US" & phase == "P4") %>%
group_by(group) %>%
summarise(value = sum(value)) %>%
ggplot(aes(x = group, y = value, fill = group)) +
geom_col()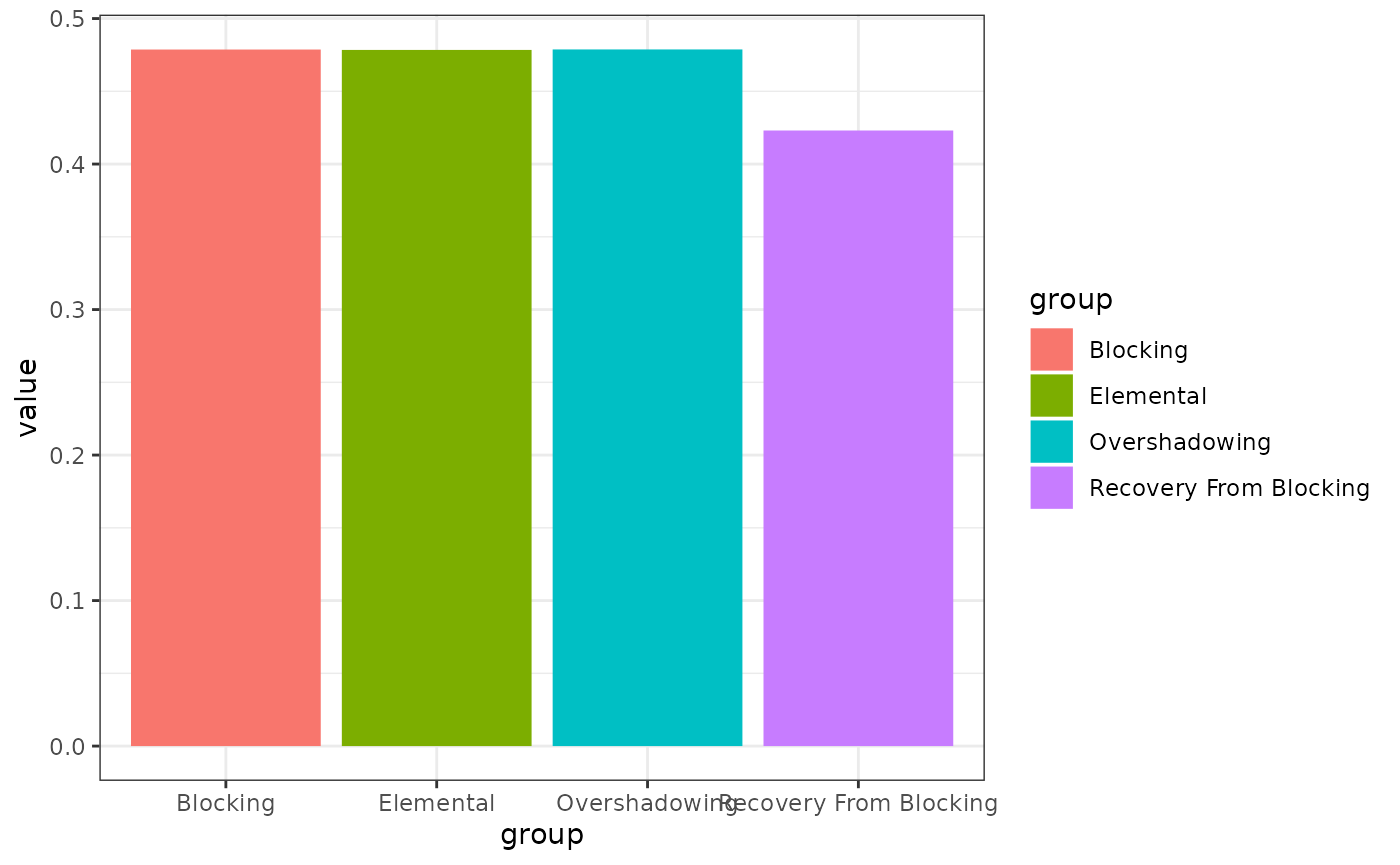
# not even close
# perhaps not enough information to do the simulation?Unequal associative changes
mod <- "SM2007"
# Gotta love a Miller design
des <- data.frame(
Group = "Unequal",
P1 = "100(ctx)X>(US)/100(ctx)XB/100(ctx)XD/100(ctx)A>(US)/100(ctx)C>(US)/1500(ctx)",
R1 = FALSE,
P2 = "3(ctx)AB>(US)/9(ctx)",
R2 = FALSE,
P3 = c("1#(ctx)AD/1#(ctx)BC"),
R3 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .35, .35, .35, .35, .50)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
m <- run_experiment(des,
model = mod, parameters = pars
) # takes a bit
# Fig 14
results(m)$relacts %>%
filter(s2 == "US" & phase == "P3") %>%
mutate(trial_type = ifelse(grepl("AD", trial_type), "Test AD", "Test BC")) %>%
group_by(trial_type) %>%
summarise(value = sum(value)) %>%
ggplot(aes(x = trial_type, y = value)) +
geom_point()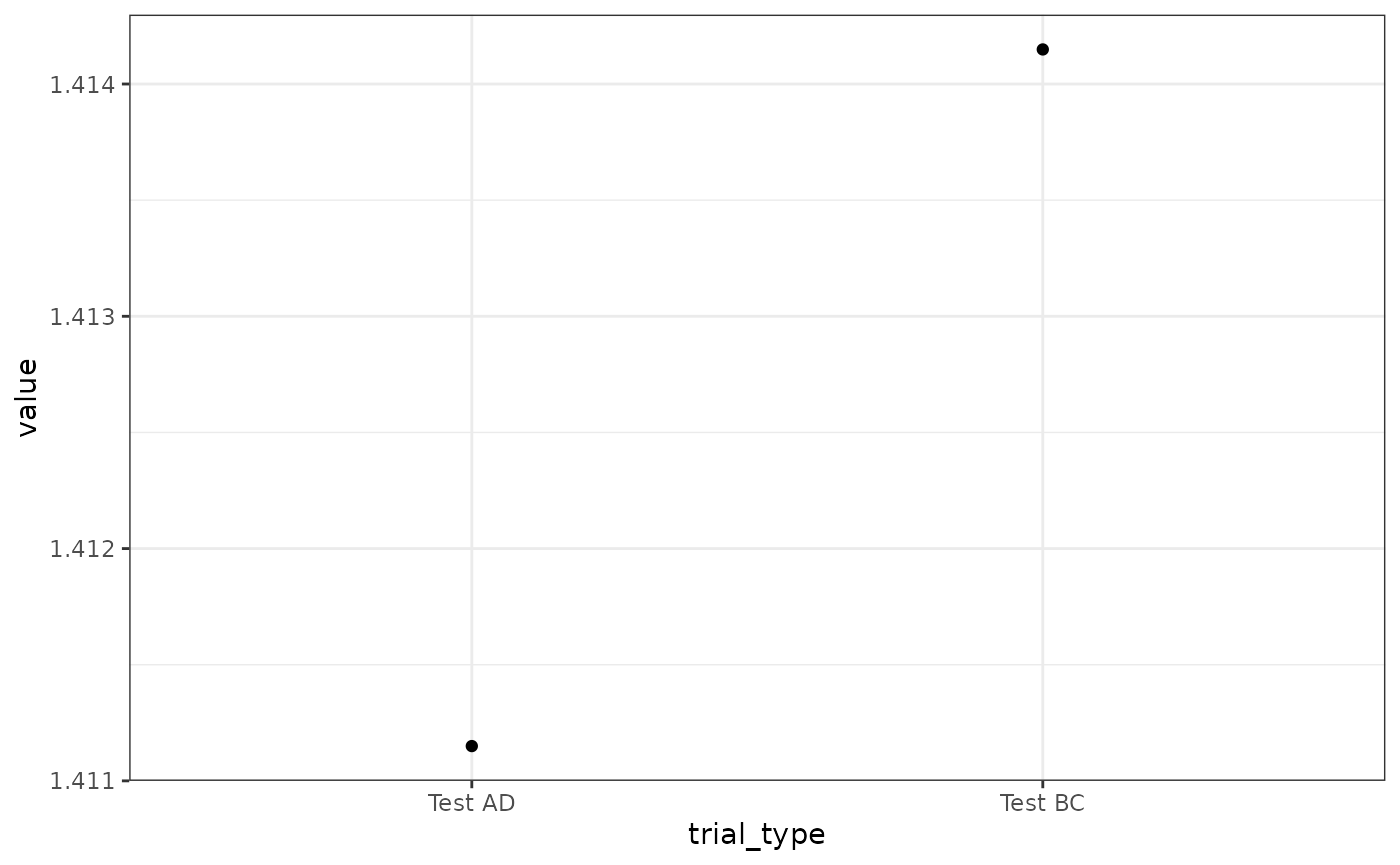
# effect in the right direction,
# but, difference is slightly bigger hereCS Preexposure
des <- data.frame(
Group = "CSPreexposure",
P1 = "66(ctx)P/198(ctx)",
R1 = FALSE,
P2 = "6(ctx)P>(US)/6(ctx)N>(US)/36(ctx)",
R2 = FALSE
)
pars <- get_parameters(des, model = mod)
pars$alphas[] <- c(.35, .15, .35, .50)
pars$omegas[] <- .22
pars$gammas[] <- .56
pars$rhos[] <- 1.6
pars$taus[] <- .46
m <- run_experiment(des,
model = mod, parameters = pars
)
# Fig 14
results(m)$relacts %>%
filter(s2 == "US" & phase == "P2" & trial_type != "(ctx)") %>%
mutate(trial = ceiling(trial / block_size)) %>%
mutate(trial_type = ifelse(grepl("P", trial_type), "Preexposed", "Novel")) %>%
group_by(trial, trial_type) %>%
summarise(value = sum(value)) %>%
ggplot(aes(x = trial, y = value, colour = trial_type)) +
geom_point() +
geom_line()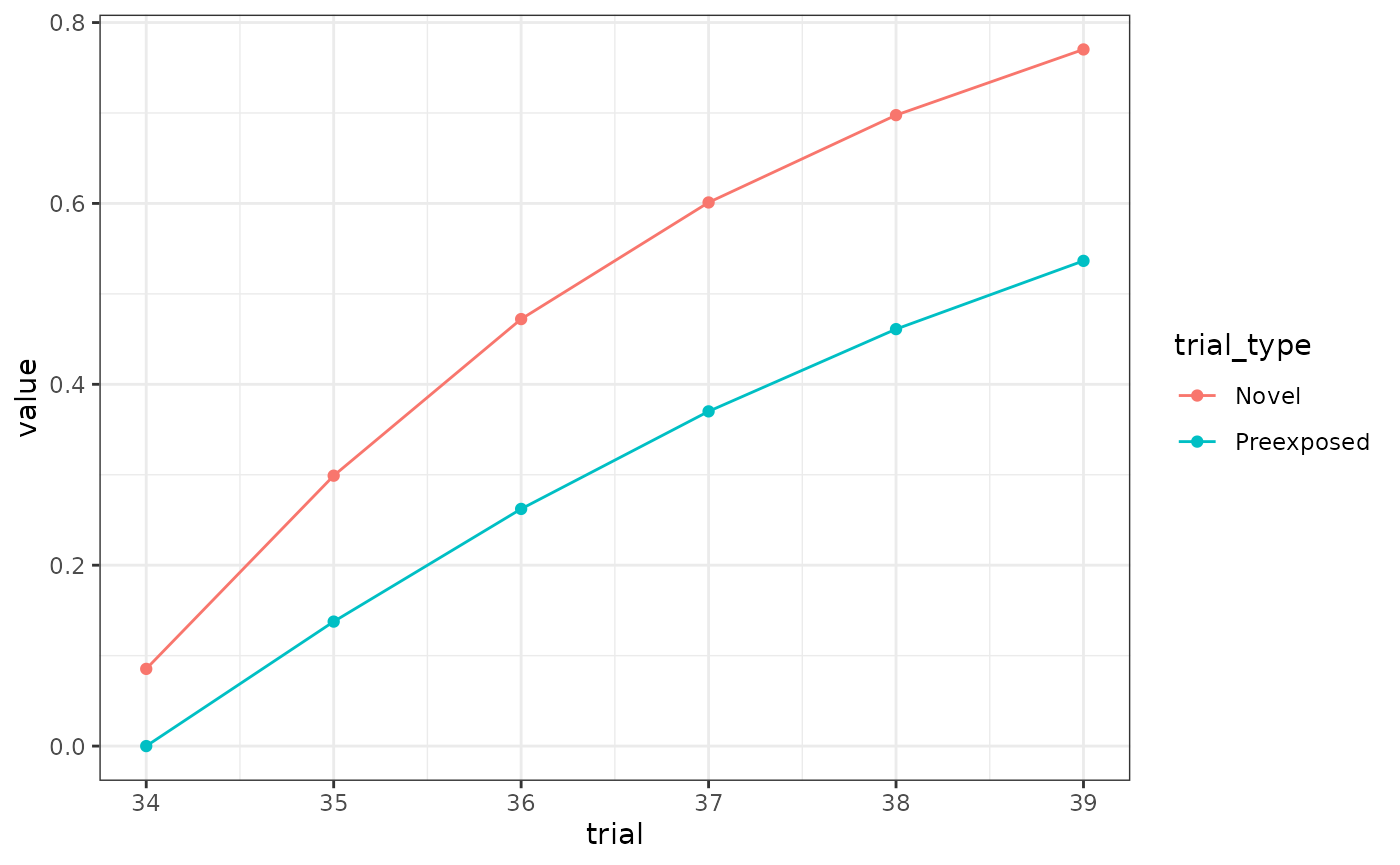
# fairly close in terms of the difference, but scale is offReferences
Witnauer, J. E., Wojick, B. M., Polack, C. W., & Miller, R. R.
(2012). Performance factors in associative learning:
Assessment of the sometimes competing retrieval model.
Learning & Behavior, 40, 347–366. https://doi.org/10.3758/s13420-012-0086-2